Basic Statistics
Summary
The module collects basic information for the sequencing reads and evaluate the reference assemblies using QUAST, a genome assembly evaluation tool.
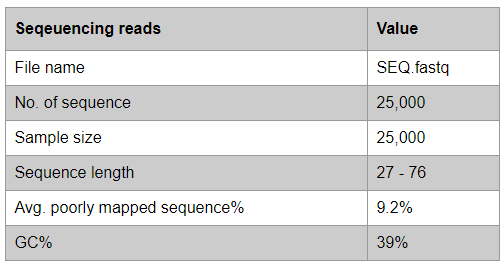
Sequencing reads
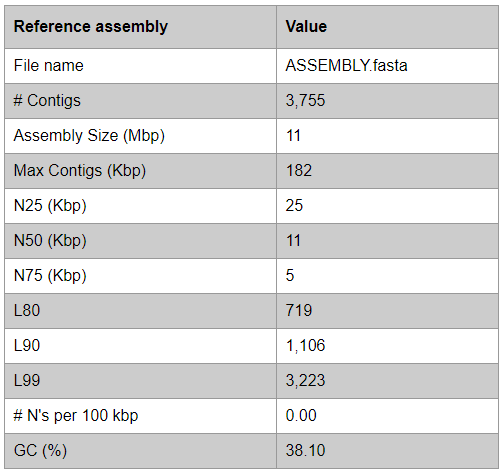
Assembly evaluation
QUAST command:
python3 SQUAT/quast/quast.py <assembly> -o <output_dir> --min-contig 200 -t <num_threads> -s
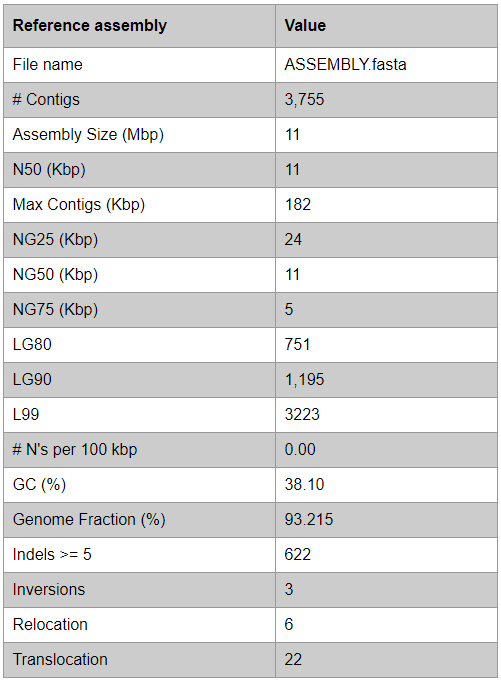
Assembly evaluation (GAGE mode)
Note that if GAGE mode is activated in QUAST, statistics such as NX and LX will be replaced by NGX and LGX in the table.
QUAST command:
python3 SQUAT/quast/quast.py <assembly> -o <output_dir> --min-contig 200 -t <num_threads> -s -R <reference genome> --gage