Read Labeling
Summary
In this section, we go through each step of read label tagging. Based on the discoveries from BWA mapping, we classify sequencing reads into seven categories.
BWA Mapping
SQUAT maps sequencing reads against their assemblies using two different algorithms: BWA-MEM and BWA-backtrack. The former algorithm performs local alignment with multiple alignments for different part of a query sequence, while the latter is more suitable for short reads and tries to map the whole sequence. For more explanation, see the manual of BWA.
Reading Labeling Workflow
The basic workflow looks as following,
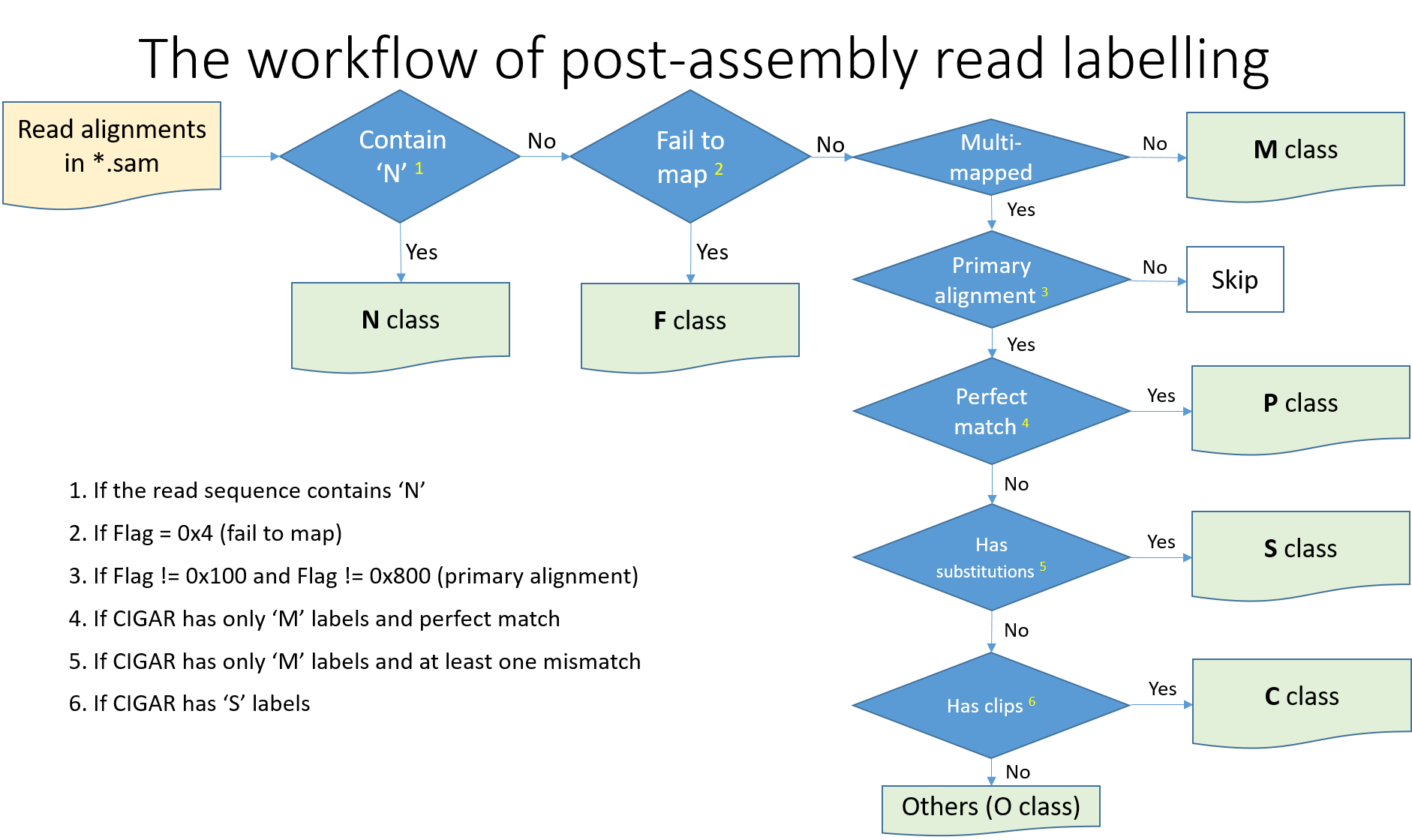
First, we label the sequences containing N to the N class. Next, based on the Flag and MAP fields in SAM file, reads indicated as unmapped and multi-mapped are assigned to F and M class respectively. The rest of reads are uniquely-mapped. They an be further divided into P(Perfect), S(substitution errors), C(contain clips), and O(other errors) class according to the CIGAR field of their primary alignment.
| Label | Description |
|---|---|
| P | Perfectly-matched reads |
| S | Reads with substitution errors |
| C | Reads that contain clips |
| O | Reads with other errors |
| M | Multi-mapped reads |
| F | Unmapped reads |
| N | Reads that contain N |
For detailed specification of SAM file, see SAM Spec.
Label Distribution
Table
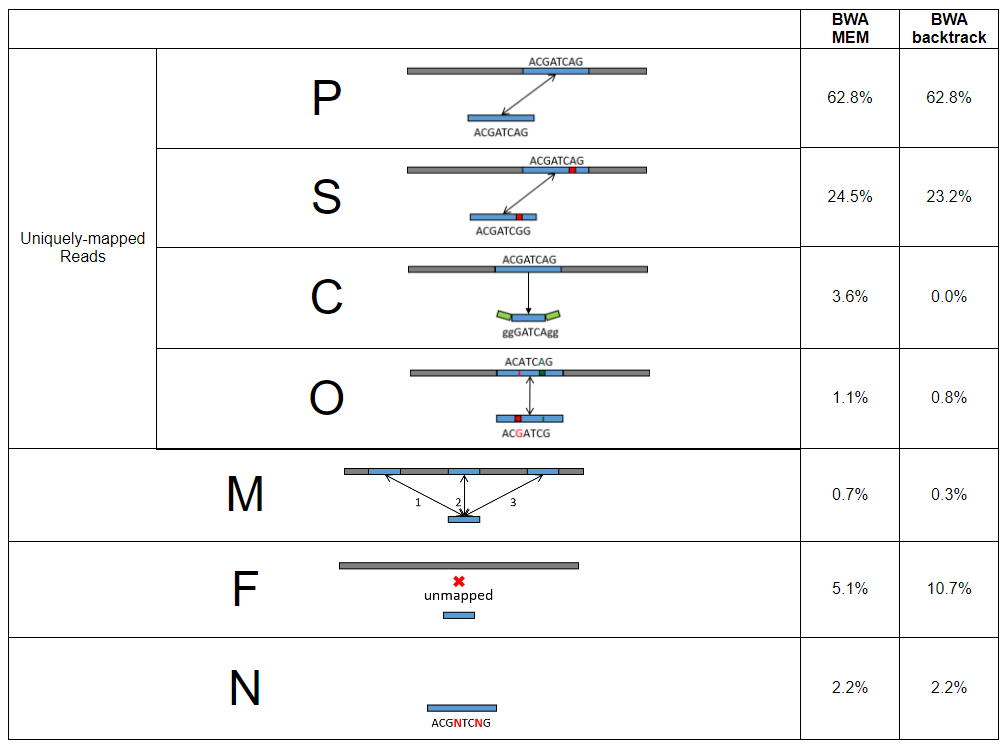
For each strategy SQUAT adopts for alignment, it lists the label distribution with an icon to represent each read class in a table.
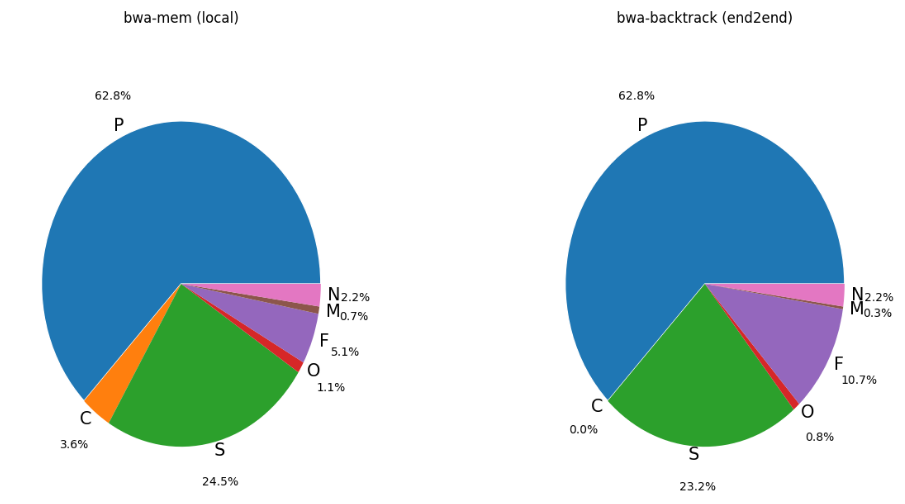
Piechart
In addition, we use a piechart to get a better sense about the label composition of data.
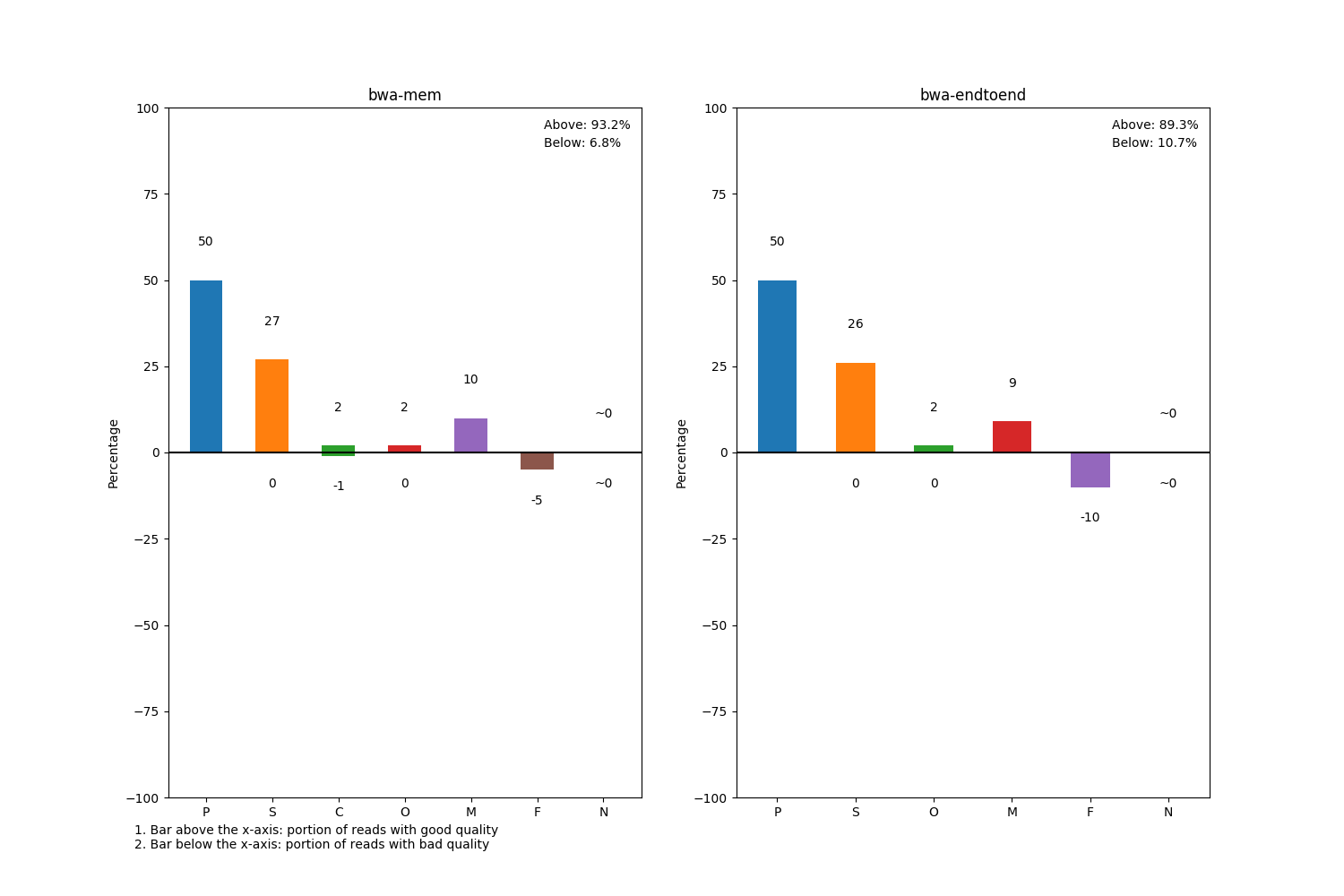
Barchart
Lastly, we plot a barchart to display the percentage of poorly-mapped reads for each read class. Reads in P and M class are always considered high quality while unmapped (F) reads will always be of poor quality. For reads in S, C, O and N class, we specify certain thresholds to determine the sequencing quality of each read. For threshold definition, see mismatch, clip and N ratio